Rows: 20,293
Columns: 78
$ ID <int> 51624, 51625, 51626, 51627, 51628, 51629, 51630, 5163…
$ SurveyYr <fct> 2009_10, 2009_10, 2009_10, 2009_10, 2009_10, 2009_10,…
$ Gender <fct> male, male, male, male, female, male, female, female,…
$ Age <int> 34, 4, 16, 10, 60, 26, 49, 1, 10, 80, 10, 80, 4, 35, …
$ AgeMonths <int> 409, 49, 202, 131, 722, 313, 596, 12, 124, NA, 121, N…
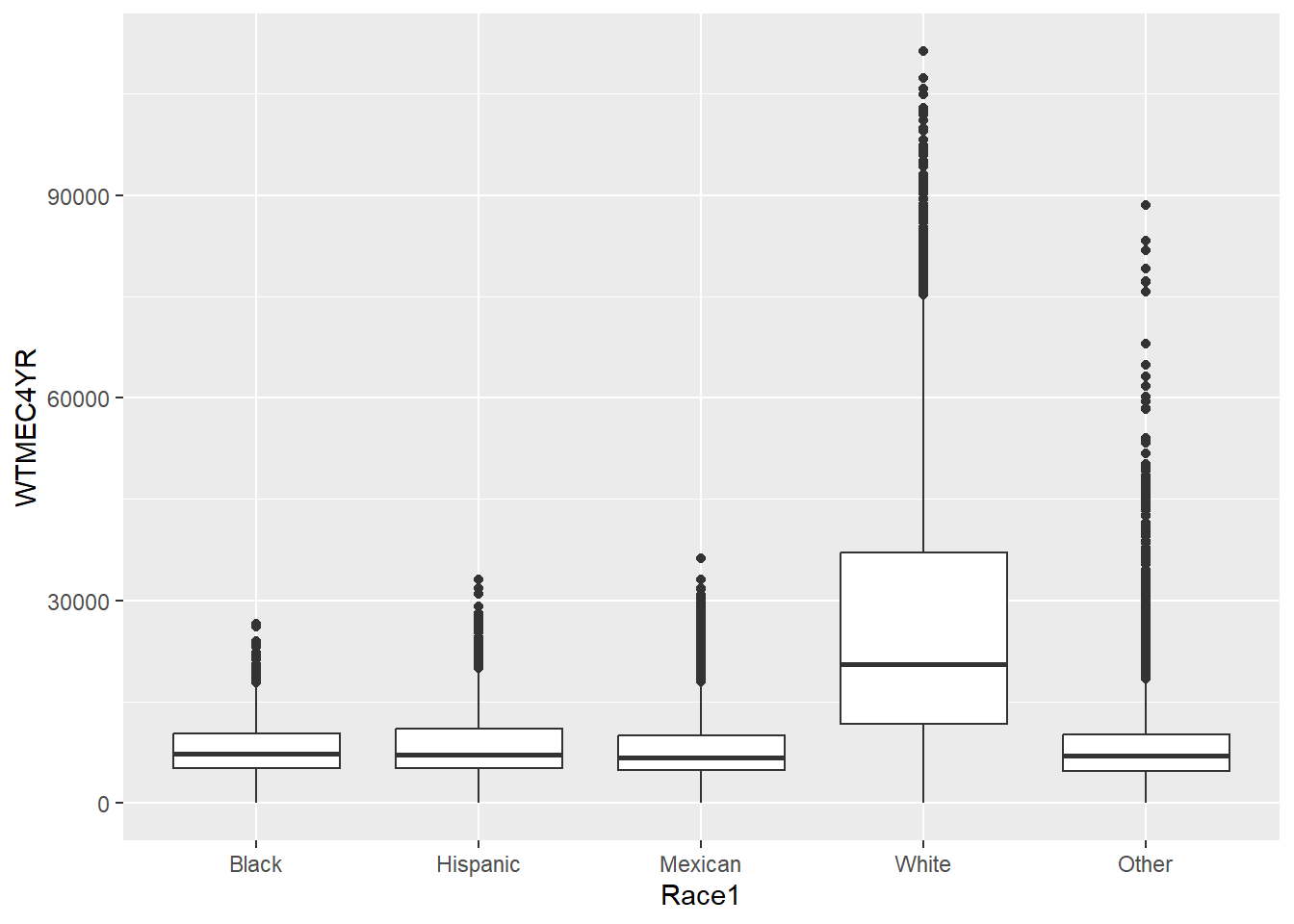
$ Race1 <fct> White, Other, Black, Black, Black, Mexican, White, Wh…
$ Race3 <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ Education <fct> High School, NA, NA, NA, High School, 9 - 11th Grade,…
$ MaritalStatus <fct> Married, NA, NA, NA, Widowed, Married, LivePartner, N…
$ HHIncome <fct> 25000-34999, 20000-24999, 45000-54999, 20000-24999, 1…
$ HHIncomeMid <int> 30000, 22500, 50000, 22500, 12500, 30000, 40000, 4000…
$ Poverty <dbl> 1.36, 1.07, 2.27, 0.81, 0.69, 1.01, 1.91, 1.36, 2.68,…
$ HomeRooms <int> 6, 9, 5, 6, 6, 4, 5, 5, 7, 4, 5, 5, 7, NA, 6, 6, 5, 6…
$ HomeOwn <fct> Own, Own, Own, Rent, Rent, Rent, Rent, Rent, Own, Own…
$ Work <fct> NotWorking, NA, NotWorking, NA, NotWorking, Working, …
$ Weight <dbl> 87.4, 17.0, 72.3, 39.8, 116.8, 97.6, 86.7, 9.4, 26.0,…
$ Length <dbl> NA, NA, NA, NA, NA, NA, NA, 75.7, NA, NA, NA, NA, NA,…
$ HeadCirc <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ Height <dbl> 164.7, 105.4, 181.3, 147.8, 166.0, 173.0, 168.4, NA, …
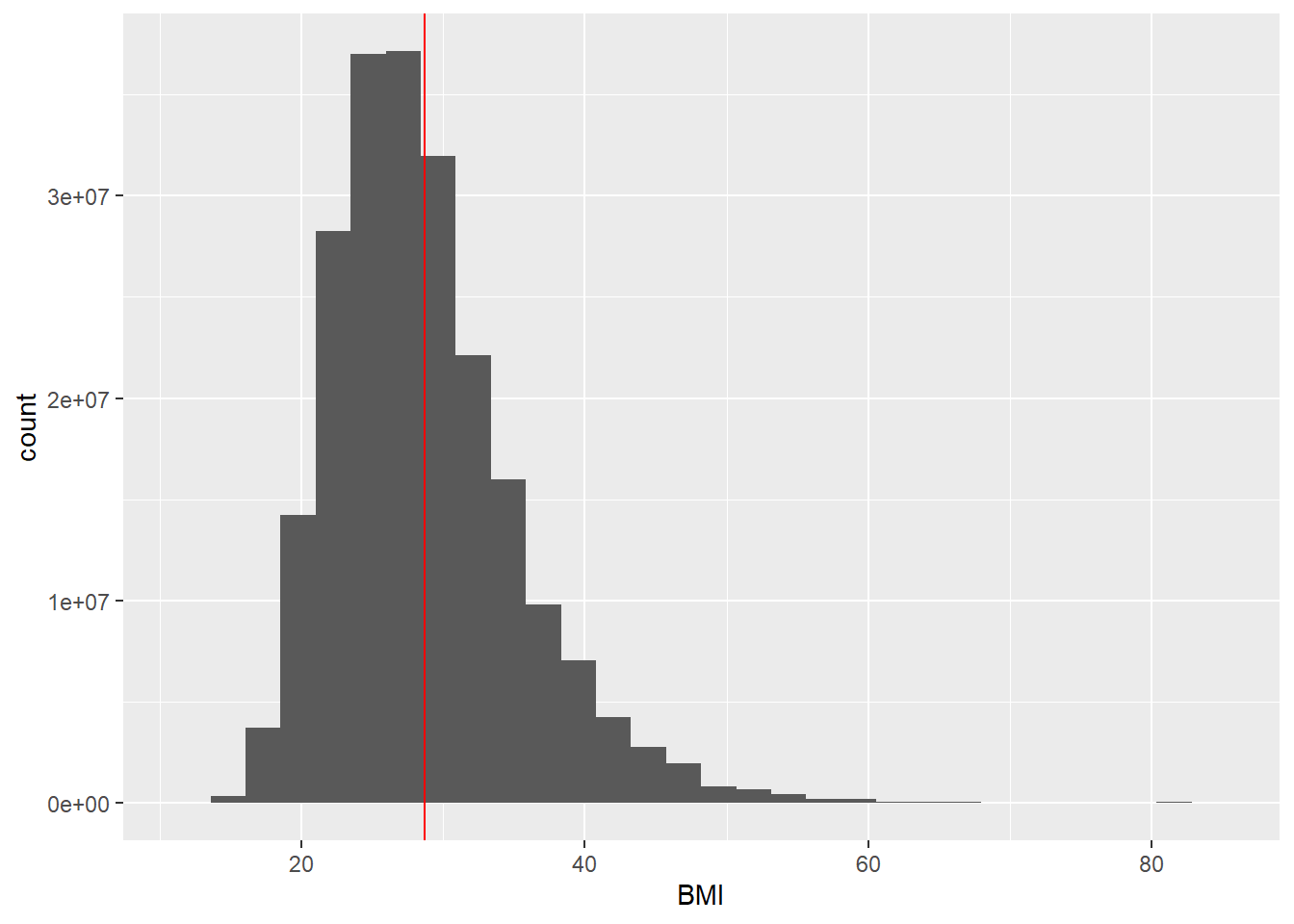
$ BMI <dbl> 32.22, 15.30, 22.00, 18.22, 42.39, 32.61, 30.57, NA, …
$ BMICatUnder20yrs <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ BMI_WHO <fct> 30.0_plus, 12.0_18.5, 18.5_to_24.9, 12.0_18.5, 30.0_p…
$ Pulse <int> 70, NA, 68, 68, 72, 72, 86, NA, 70, 88, 84, 54, NA, N…
$ BPSysAve <int> 113, NA, 109, 93, 150, 104, 112, NA, 108, 139, 94, 12…
$ BPDiaAve <int> 85, NA, 59, 41, 68, 49, 75, NA, 53, 43, 45, 60, NA, N…
$ BPSys1 <int> 114, NA, 112, 92, 154, 102, 118, NA, 106, 142, 94, 12…
$ BPDia1 <int> 88, NA, 62, 36, 70, 50, 82, NA, 60, 62, 38, 62, NA, N…
$ BPSys2 <int> 114, NA, 114, 94, 150, 104, 108, NA, 106, 140, 92, 12…
$ BPDia2 <int> 88, NA, 60, 44, 68, 48, 74, NA, 50, 46, 40, 62, NA, N…
$ BPSys3 <int> 112, NA, 104, 92, 150, 104, 116, NA, 110, 138, 96, 11…
$ BPDia3 <int> 82, NA, 58, 38, 68, 50, 76, NA, 56, 40, 50, 58, NA, N…
$ Testosterone <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ DirectChol <dbl> 1.29, NA, 1.55, 1.89, 1.16, 1.16, 1.16, NA, 1.58, 1.9…
$ TotChol <dbl> 3.49, NA, 4.97, 4.16, 5.22, 4.14, 6.70, NA, 4.14, 4.7…
$ UrineVol1 <int> 352, NA, 281, 139, 30, 202, 77, NA, 39, 128, 109, 38,…
$ UrineFlow1 <dbl> NA, NA, 0.415, 1.078, 0.476, 0.563, 0.094, NA, 0.300,…
$ UrineVol2 <int> NA, NA, NA, NA, 246, NA, NA, NA, NA, NA, NA, NA, NA, …
$ UrineFlow2 <dbl> NA, NA, NA, NA, 2.51, NA, NA, NA, NA, NA, NA, NA, NA,…
$ Diabetes <fct> No, No, No, No, Yes, No, No, No, No, No, No, Yes, No,…
$ DiabetesAge <int> NA, NA, NA, NA, 56, NA, NA, NA, NA, NA, NA, 70, NA, N…
$ HealthGen <fct> Good, NA, Vgood, NA, Fair, Good, Good, NA, NA, Excell…
$ DaysPhysHlthBad <int> 0, NA, 2, NA, 20, 2, 0, NA, NA, 0, NA, 0, NA, NA, NA,…
$ DaysMentHlthBad <int> 15, NA, 0, NA, 25, 14, 10, NA, NA, 0, NA, 0, NA, NA, …
$ LittleInterest <fct> Most, NA, NA, NA, Most, None, Several, NA, NA, None, …
$ Depressed <fct> Several, NA, NA, NA, Most, Most, Several, NA, NA, Non…
$ nPregnancies <int> NA, NA, NA, NA, 1, NA, 2, NA, NA, NA, NA, NA, NA, NA,…
$ nBabies <int> NA, NA, NA, NA, 1, NA, 2, NA, NA, NA, NA, NA, NA, NA,…
$ Age1stBaby <int> NA, NA, NA, NA, NA, NA, 27, NA, NA, NA, NA, NA, NA, N…
$ SleepHrsNight <int> 4, NA, 8, NA, 4, 4, 8, NA, NA, 6, NA, 9, NA, 7, NA, N…
$ SleepTrouble <fct> Yes, NA, No, NA, No, No, Yes, NA, NA, No, NA, No, NA,…
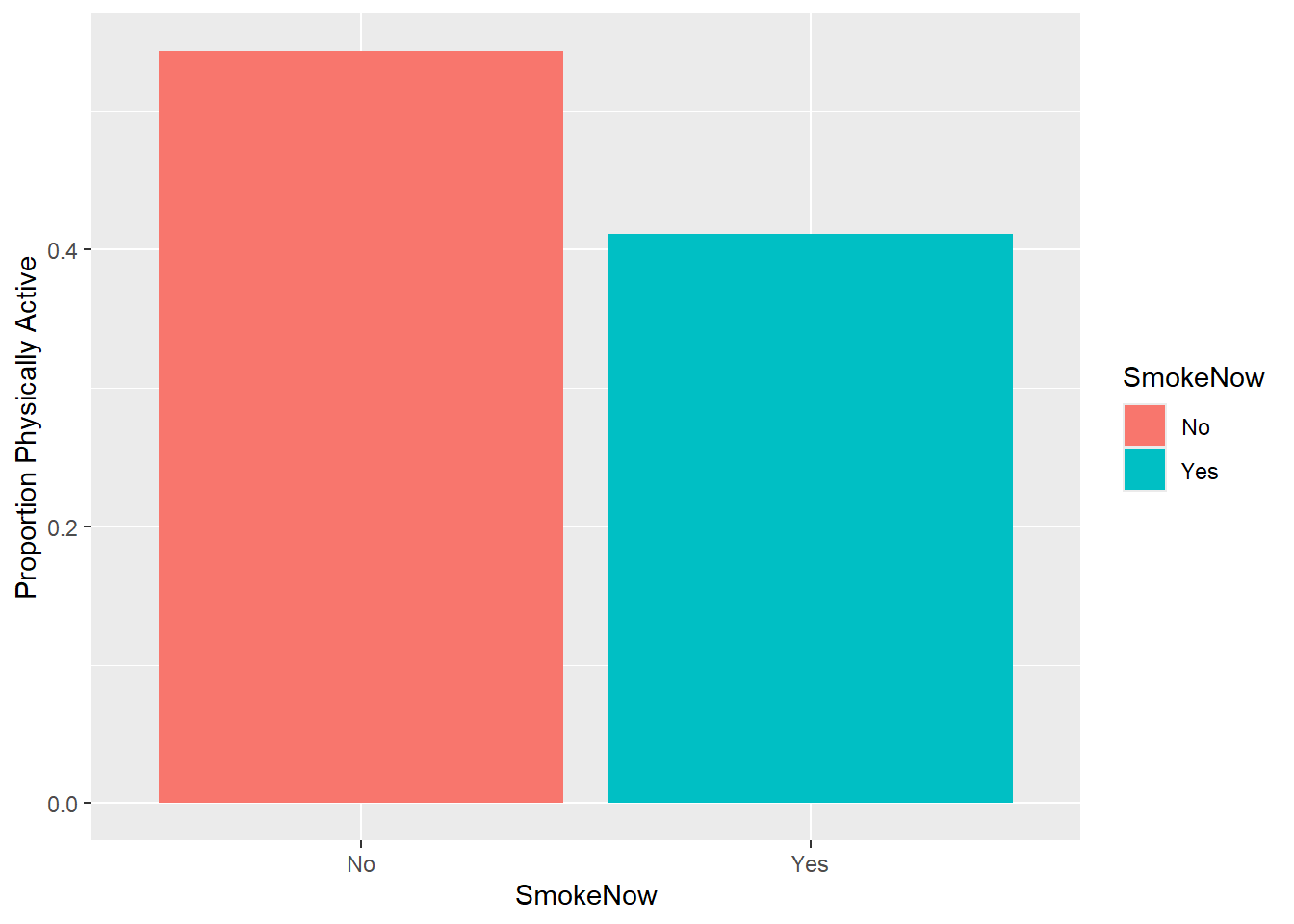
$ PhysActive <fct> No, NA, Yes, NA, No, Yes, No, NA, NA, Yes, NA, No, NA…
$ PhysActiveDays <int> NA, NA, 5, NA, NA, 2, NA, NA, NA, 4, NA, NA, NA, NA, …
$ TVHrsDay <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ CompHrsDay <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ TVHrsDayChild <int> NA, 4, NA, 1, NA, NA, NA, NA, 1, NA, 3, NA, 2, NA, 5,…
$ CompHrsDayChild <int> NA, 1, NA, 1, NA, NA, NA, NA, 0, NA, 0, NA, 1, NA, 0,…
$ Alcohol12PlusYr <fct> Yes, NA, NA, NA, No, Yes, Yes, NA, NA, Yes, NA, No, N…
$ AlcoholDay <int> NA, NA, NA, NA, NA, 19, 2, NA, NA, 1, NA, NA, NA, NA,…
$ AlcoholYear <int> 0, NA, NA, NA, 0, 48, 20, NA, NA, 52, NA, 0, NA, NA, …
$ SmokeNow <fct> No, NA, NA, NA, Yes, No, Yes, NA, NA, No, NA, No, NA,…
$ Smoke100 <fct> Yes, NA, NA, NA, Yes, Yes, Yes, NA, NA, Yes, NA, Yes,…
$ SmokeAge <int> 18, NA, NA, NA, 16, 15, 38, NA, NA, 16, NA, 21, NA, N…
$ Marijuana <fct> Yes, NA, NA, NA, NA, Yes, Yes, NA, NA, NA, NA, NA, NA…
$ AgeFirstMarij <int> 17, NA, NA, NA, NA, 10, 18, NA, NA, NA, NA, NA, NA, N…
$ RegularMarij <fct> No, NA, NA, NA, NA, Yes, No, NA, NA, NA, NA, NA, NA, …
$ AgeRegMarij <int> NA, NA, NA, NA, NA, 12, NA, NA, NA, NA, NA, NA, NA, N…
$ HardDrugs <fct> Yes, NA, NA, NA, No, Yes, Yes, NA, NA, NA, NA, NA, NA…
$ SexEver <fct> Yes, NA, NA, NA, Yes, Yes, Yes, NA, NA, NA, NA, NA, N…
$ SexAge <int> 16, NA, NA, NA, 15, 9, 12, NA, NA, NA, NA, NA, NA, NA…
$ SexNumPartnLife <int> 8, NA, NA, NA, 4, 10, 10, NA, NA, NA, NA, NA, NA, NA,…
$ SexNumPartYear <int> 1, NA, NA, NA, NA, 1, 1, NA, NA, NA, NA, NA, NA, NA, …
$ SameSex <fct> No, NA, NA, NA, No, No, Yes, NA, NA, NA, NA, NA, NA, …
$ SexOrientation <fct> Heterosexual, NA, NA, NA, NA, Heterosexual, Heterosex…
$ WTINT2YR <dbl> 80100.544, 53901.104, 13953.078, 11664.899, 20090.339…
$ WTMEC2YR <dbl> 81528.772, 56995.035, 14509.279, 12041.635, 21000.339…
$ SDMVPSU <int> 1, 2, 1, 2, 2, 1, 2, 2, 2, 1, 1, 1, 2, 2, 1, 1, 1, 1,…
$ SDMVSTRA <int> 83, 79, 84, 86, 75, 88, 85, 86, 88, 77, 86, 79, 84, 7…
$ PregnantNow <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, U…